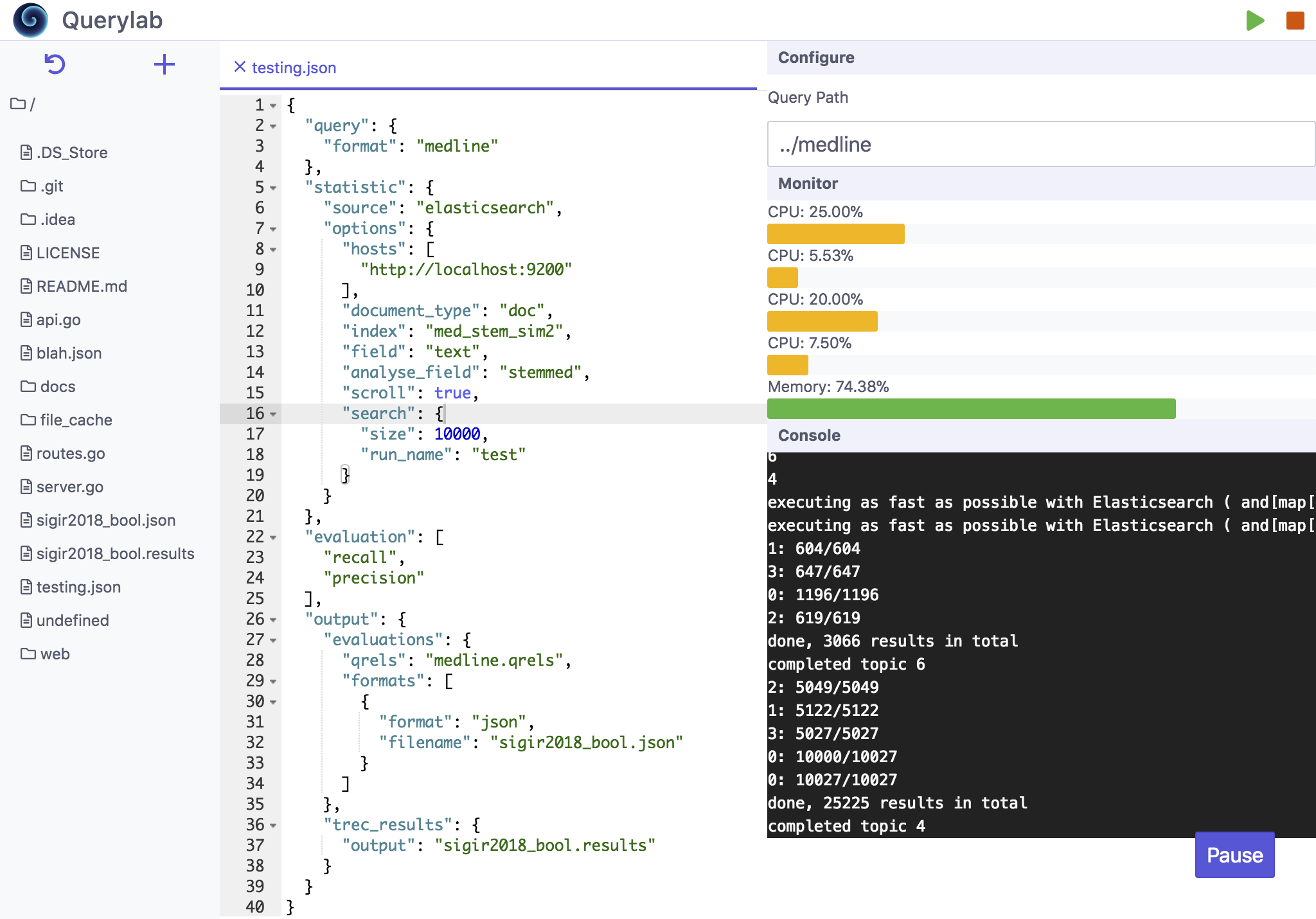

QueryLab is a framework for constructing and executing information retrieval experiment pipelines. The framework as a whole is built primarily for domain specific applications such as medical literature search for systematic reviews, or finding factually or legally applicable case law in the legal domain; however it can also be used for more general tasks. There are a number of pre-implemented components that enable common information retrieval experiments such as ad-hoc retrieval or query analysis through query performance predictors. In addition, this collection of tools seeks to be user friendly, well documented, and easily extendible. Finally, the entire pipeline can be distributed as a single binary with no dependencies, ready to use with a simple domain specific language (DSL) for constructing pipelines.
The syntax of Querylab pipelines look like the following:
{
"query": {
"source": "medline"
},
"statistic": {
"source": "elasticsearch"
},
"measurements": [
"keyword_count",
"term_count",
"avg_idf",
"avg_ictf"
],
"preprocess": [
"lowercase"
],
"output": [
{
"format": "json",
"filename": "analysis.json"
},
{
"format": "csv",
"filename": "analysis.csv"
}
]
}
More examples and documentation for how to construct pipelines can be found at GitHub.
The Querylab interface comprises four major aspects. Each of these aspects have their own command line utilities and GitHub repositories and can in fact work independently of each other. Querylab is the merging of these four components into one IDE-like interface.
The common query representation module is used to simplify experiments in the rest of the pipeline. It forms the basis for how queries are represented and specifies how they can be transformed. The representation is similar to that of the Elasticsearch DSL. There are two possible representations of a query: the first is a keyword which is similar to what is seen in web search — a string of characters restricted to some fields; the second is a Boolean query — which combines keyword queries with logical operators. A CQR query in human-readable notation takes the form of JSON.
https://github.com/hscells/cqr
When replicating a domain-specific IR task, it is often required to use a specific query language. These queries can either be manually translated by hand or automatically by a parser/compiler into a target query language for use in an IR system. The transmute library and command-line tool is a parser/compiler for queries from one query language to another. Currently, transmute can transform Medline and PubMed queries, and CQR queries into Elasticsearch queries (the Terrier query language is currently being implemented).</p>
https://github.com/hscells/transmute
The groove library provides abstractions and implementations for performing IR experiments. A groove pipeline comprises a query source (the format of the queries), a statistic source (a source for computing IR statistics; i.e. a search engine such as Elasticsearch or Terrier), preprocessing steps (e.g., lowercase, stemming, stopword removal), any measurements to make (e.g., query performance predictors, retrieval results, evaluation criteria), and any output formats (e.g., JSON, csv). Each component in the pipeline is extendible and well documented.
Currently, groove can load Medline and PubMed queries (query languages commonly used in systematic reviews) via transmute, and LexesNexes queries (a query language commonly used in legal IR) via an implementation integrated into groove.
https://github.com/hscells/groove
The boogie command-line utility provides higher level access to groove. It allows for the specification of a pipeline to be written in a domain specific language (DSL) that groove can then execute. At a high level, the purpose of this pipeline is to compute measurements for MEDLINE queries between a number of query performance predictors and the performance of the actual effectiveness of the queries in terms of precision and recall.
At a deeper level, reading the pipeline from top to bottom: the type of queries are specified (MEDLINE), the source of statistical information (i.e. search engine) is configured to point to an Elasticsearch instance, the list of query performance predictors measurements are listed followed by the evaluation measures to record, and, finally, the last item specifies how the results of the pipeline should be output and in which formats (the trec_eval-style results file is also output so as to record the retrieval results.
https://github.com/hscells/boogie
If you happen to use this work, please cite it as:
@inproceedings{scells2017querylab,
Author = {Scells, Harrisen and Zuccon, Guido and Locke, Daniel},
Booktitle = {Proceedings of the 41st international ACM SIGIR conference on Research and development in Information Retrieval},
Organization = {ACM},
Title = {An Information Retrieval Experiment Framework for Domain Specific Applications},
Year = {2018}
}
For support, please open an issue on the respective GitHub repositories, or on the Querylab Github repository, at https://github.com/ielab/querylab.
Last update at 2019-03-27 01:55:28 +0000